Contact
Positions
Professor
- Organization:
- West Virginia University School of Medicine
- Department:
- Biochemistry and Molecular Medicine
- Classification:
- Faculty
Professor
- Organization:
- West Virginia University School of Medicine
- Department:
- Department of Neuroscience
- Classification:
- Faculty
Member
- Organization:
- West Virginia University WVU Cancer Institute
- Department:
- WVU Cancer Institute Research Programs
- Classification:
- Faculty
Professor
- Organization:
- West Virginia University School of Medicine
- Department:
- Rockefeller Neuroscience Institute (SOM)
- Classification:
- Faculty
Publications
Thomas T, Salcedo-Tacuma D, Smith DM. Structure, Function, and Allosteric Regulation of the 20S Proteasome by the 11S/PA28 Family of Proteasome Activators. Biomolecules. 2023 Aug 29;13(9). doi: 10.3390/biom13091326. Review. PubMed PMID: 37759726; PubMed Central PMCID: PMC10526260.
Chuah, J.J.Y., Rexroad, M.S. & Smith, D.M. High resolution structures define divergent and convergent mechanisms of archaeal proteasome activation. Commun Biol 6, 733 (2023). https://doi.org/10.1038/s42003-023-05123-3
Chuah, J.J.Y., Thibaudeau, T.A. & Smith, D.M. Minimal mechanistic component of HbYX-dependent proteasome activation that reverses impairment by neurodegenerative-associated oligomers. Commun Biol 6, 725 (2023). https://doi.org/10.1038/s42003-023-05082-9
Anderson R, Bradley T, Smith DM. Hyperactivation of the proteasome in C. elegans protects against proteotoxic stress and extends lifespan. J Biol Chem. 2022 Aug 23;298(10):102415. doi: 10.1016/j.jbc.2022.102415. PubMed PMID: 36007615; PubMed Central PMCID: PMC9486566.
Thomas, TA and Smith DM. Proteasome activator 28γ (PA28γ) allosterically activates trypsin-like proteolysis by binding to the α-ring of the 20S proteasome. Journal of Biological Chemistry (2022) Vol. 298, Issue 8, 102140. PMID: 35714770
Pilkington AW 4th, Schupp J, Nyman M, Valentine SJ, Smith DM, Legleiter J. Acetylation of Aβ40 Alters Aggregation in the Presence and Absence of Lipid Membranes. ACS Chem Neurosci. 2020 Jan 15;11(2):146-161. doi: 10.1021/acschemneuro.9b00483. Epub 2019 Dec 27. PubMed PMID: 31834770; PubMed Central PMCID: PMC7477891
DeVallance E, Branyan KW, Lemaster KC, Anderson R, Marshall KL, Olfert IM, Smith DM, Kelley EE, Bryner RW, Frisbee JC, Chantler PD. Exercise training prevents the perivascular adipose tissue-induced aortic dysfunction with metabolic syndrome. Redox Biol. 2019 Sep;26:101285. doi: 10.1016/j.redox.2019.101285. Epub 2019 Jul 26. PubMed PMID: 31374361; PubMed Central PMCID: PMC6669320
Thibaudeau TA & Smith DM. "A practical Review of Proteasome Pharmacology" Pharmacological Reviews 71:170-197, April 2019
Brooks C., Snoberger A., Belcastro M., Murphy J., Kisselev O.G., Smith D.M. and Sokolov M., , “Archaeal Unfoldase Counteracts Protein Misfolding Retinopathy in Mice” Journal of Neuroscience 38 (33) 7248-7254 (2018)
Snoberger, A., Brettrager, E & Smith DM (2018). "Conformational switching in the coiled-coil domains of a proteasomal ATPase regulates substrate processing." Nat Commun 9(1): 2374.
DeVallance E, Branyan KW, Lemaster K, Olfert IM, Smith DM, Pistilli EE, Frisbee JC, Chantler PD. "Aortic dysfunction in metabolic syndrome mediated by perivascular adipose tissue TNFα- and NOX2-dependent pathway." Exp Physiol. 2018 Apr 1;103(4):590-603. doi: 10.1113/EP086818. Epub 2018 Feb 28.
Thibaudeau TA, Anderson RT, Smith DM. "A common mechanism of proteasome impairment by neurodegenerative disease-associated oligomers." Nature Communications. 2018 Mar 15;9(1):1097. doi: 10.1038/s41467-018-03509-0. PubMed PMID: 29545515
Snoberger A, Anderson RT, Smith DM "The Proteasomal ATPases Use a Slow but Highly Processive Strategy to Unfold Proteins." Front Mol Biosci. 2017 Apr 4;4:18. doi: 10.3389/fmolb.2017.00018. eCollection 2017.
Kim YC, Snoberger A, Schupp J, Smith DM. "ATP binding to neighbouring subunits and intersubunit allosteric coupling underlie proteasomal ATPase function.". Nature Communications 2015; 6:8520. NIHMSID: NIHMS719879 PubMed [journal] PMID:26465836, PMCID: PMC4608255
Deriziotis P, André R, Smith DM, Goold R, Kinghorn KJ, Kristiansen M, Nathan JA, Rosenzweig R, Krutauz D, Glickman MH, Collinge J, Goldberg AL, Tabrizi SJ. "Misfolded PrP impairs the UPS by interaction with the 20S proteasome and inhibition of substrate entry". The EMBO journal. 2011; 30(15):3065-77. PubMed [journal] PMID: 21743439, PMCID: PMC3160194
Bader M, Benjamin S, Wapinski OL, Smith DM, Goldberg AL, Steller H. "A conserved F box regulatory complex controls proteasome activity in Drosophila". Cell. 2011; 145(3):371-82. NIHMSID: NIHMS293104 PubMed [journal] PMID: 21529711, PMCID:PMC3108249
Smith DM, Fraga H, Reis C, Kafri G, Goldberg AL. "ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle". Cell. 2011; 144(4):526-38. NIHMSID: NIHMS272218 PubMed [journal] PMID: 21335235, PMCID: PMC3063399
Yu Y, Smith DM, Kim HM, Rodriguez V, Goldberg AL, Cheng Y. "Interactions of PAN's C-termini with archaeal 20S proteasome and implications for the eukaryotic proteasome-ATPase interactions". The EMBO journal. 2010; 29(3):692-702. PubMed [journal] PMID: 20019667, PMCID: PMC2830694
Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. "Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases". Molecular cell. 2008; 30(3):360-8. NIHMSID: NIHMS156029 PubMed [journal] PMID: 18471981, PMCID: PMC4141531
Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. "Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry". Molecular cell. 2007; 27(5):731-44. NIHMSID: NIHMS30186 PubMed [journal] PMID: 17803938, PMCID: PMC2083707
Horwitz AA, Navon A, Groll M, Smith DM, Reis C, Goldberg AL. "ATP-induced structural transitions in PAN, the proteasome-regulatory ATPase complex in Archaea". The Journal of biological chemistry. 2007; 282(31):22921-9. PubMed [journal] PMID: 17553803
Smith DM, Benaroudj N, Goldberg A. "Proteasomes and their associated ATPases: a destructive combination. Journal of structural biology". 2006; 156(1):72-83. PubMed [journal] PMID: 16919475
Lu M, Dou QP, Kitson RP, Smith DM, Goldfarb RH. "Differential effects of proteasome inhibitors on cell cycle and apoptotic pathways in human YT and Jurkat cells". Journal of cellular biochemistry. 2006; 97(1):122-34. PubMed [journal] PMID: 16173095
Smith DM, Kafri G, Cheng Y, Ng D, Walz T, Goldberg AL. "ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins". Molecular cell. 2005; 20(5):687-98. PubMed [journal] PMID: 16337593
Arbiser JL, Li XC, Hossain CF, Nagle DG, Smith DM, Miller P, Govindarajan B, DiCarlo J, Landis-Piwowar KR, Dou QP. "Naturally occurring proteasome inhibitors from mate tea (Ilex paraguayensis) serve as models for topical proteasome inhibitors". The Journal of investigative dermatology. 2005; 125(2):207-12. PubMed [journal] PMID: 16098028
Smith DM, Daniel KG, Wang Z, Guida WC, Chan TH, Dou QP. "Docking studies and model development of tea polyphenol proteasome inhibitors: applications to rational drug design". Proteins. 2004; 54(1):58-70. PubMed [journal] PMID: 14705024
Kazi A, Daniel KG, Smith DM, Kumar NB, Dou QP. "Inhibition of the proteasome activity, a novel mechanism associated with the tumor cell apoptosis-inducing ability of genistein". Biochemical pharmacology. 2003; 66(6):965-76. PubMed [journal] PMID: 12963483
Kuhn DJ, Smith DM, Pross S, Whiteside TL, Dou QP. "Overexpression of interleukin-2 receptor alpha in a human squamous cell carcinoma of the head and neck cell line is associated with increased proliferation, drug resistance, and transforming ability". Journal of cellular biochemistry. 2003; 89(4):824-36. PubMed [journal] PMID: 12858347
Dou QP, Smith DM, Daniel KG, Kazi A. "Interruption of tumor cell cycle progression through proteasome inhibition: implications for cancer therapy". Progress in cell cycle research. 2003; 5:441-6. PubMed [journal] PMID: 14593738
Kazi A, Smith DM, Daniel K, Zhong S, Gupta P, Bosley ME, Dou QP. "Potential molecular targets of tea polyphenols in human tumor cells: significance in cancer prevention". In vivo (Athens, Greece). 2002; 16(6):397-403. PubMed [journal] PMID:12494882
Kazi A, Smith DM, Zhong Q, Dou QP. "Inhibition of bcl-x(l) phosphorylation by tea polyphenols or epigallocatechin-3-gallate is associated with prostate cancer cell apoptosis". Molecular pharmacology. 2002; 62(4):765-71. PubMed [journal] PMID:12237322
Smith DM, Wang Z, Kazi A, Li LH, Chan TH, Dou QP. "Synthetic analogs of green tea polyphenols as proteasome inhibitors". Molecular medicine (Cambridge, Mass.). 2002; 8(7):382-92. PubMed [journal] PMID: 12393936, PMCID: PMC2040000
Smith DM, Kazi A, Smith L, Long TE, Heldreth B, Turos E, Dou QP. "A novel beta-lactam antibiotic activates tumor cell apoptotic program by inducing DNA damage". Molecular pharmacology. 2002; 61(6):1348-58. PubMed [journal] PMID:12021396
Nam S, Smith DM, Dou QP. "Tannic acid potently inhibits tumor cell proteasome activity, increases p27 and Bax expression, and induces G1 arrest and apoptosis". Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2001; 10(10):1083-8. PubMed [journal] PMID: 11588135
Smith DM, Dou QP. "Green tea polyphenol epigallocatechin inhibits DNA replication and consequently induces leukemia cell apoptosis". International journal of molecular medicine. 2001; 7(6):645-52. PubMed [journal] PMID: 11351279
Nam S, Smith DM, Dou QP. "Ester bond-containing tea polyphenols potently inhibit proteasome activity in vitro and in vivo". The Journal of biological chemistry. 2001; 276(16):13322-30. PubMed [journal] PMID: 11278274
Smith DM, Gao G, Zhang X, Wang G, Dou QP. "Regulation of tumor cell apoptotic sensitivity during the cell cycle (Review)". International journal of molecular medicine. 2000; 6(5):503-7. PubMed [journal] PMID: 11029514
Research Program
Alexander B. Osborn Hematopoietic Malignancy and Transplantation Program
Research Interests
Mechanics of Proteasome Function
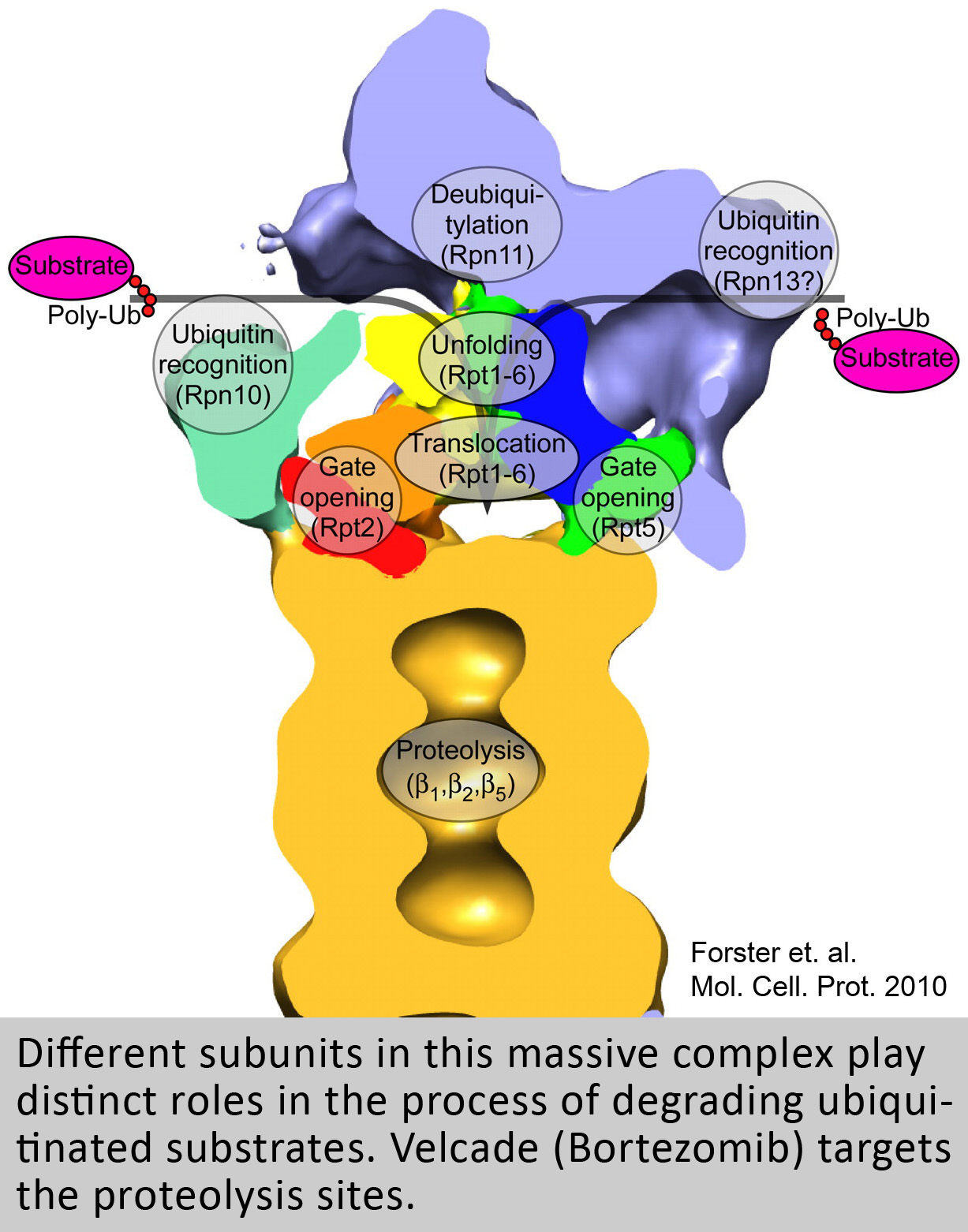
The general interest of our lab is to understand the function of the proteasome, a giant molecular machine that has the capacity to destroy nearly every protein in the cell. Despite this destructive capability the proteasome is highly selective in what it chooses to destroy. In fact, this machine is so selective and precise that it can degrade a single subunit out of a protein complex with surgical precision, or can amputate a single domain from an isolated protein. Ultimately, our lab is interested in understanding how the proteasome moves and functions at a molecular level, using an energy dependent multistep process involving 1) substrate binding, 2) unfolding, 3) translocation, 4) gate-opening and 5) destruction inside of a sequestered chamber. More specifically we have a special interest in understanding how the many different regulatory “caps” that bind to the 20S proteasome catalyze aspects of this multistep process. Because of the proteasome’s central role in regulating most cellular processes (e.g. Cell cycle, Apoptosis, transcription, receptor signaling, etc.) understanding the mechanisms that regulate its specificity is not only of biological interest, but is also highly relevant to many areas of medicine (e.g. Cancer and Neurodegenerative disease).
Animated model of how a ubiquitinated substrate is degraded by the 26S proteasome.
- The 19S binds to the 20S while hydrolyzing ATP.
- The ATPases C-terminal HbYX motif works like a key-in-a-lock to induce gate opening.
- A Ubiquitinated (orange) substrate (green) binds to the 19S.
- The substrate is unfolded and translocated through the opened gate.
- The substrate is degraded by protease sites inside the 20S chamber.
- Intact ubiquitin and small peptide products are released during this ATP-dependent process.
Animation introducing the general structure of the 26S proteasome.
(Movies were created in collaboration with, Janet Iwasa)
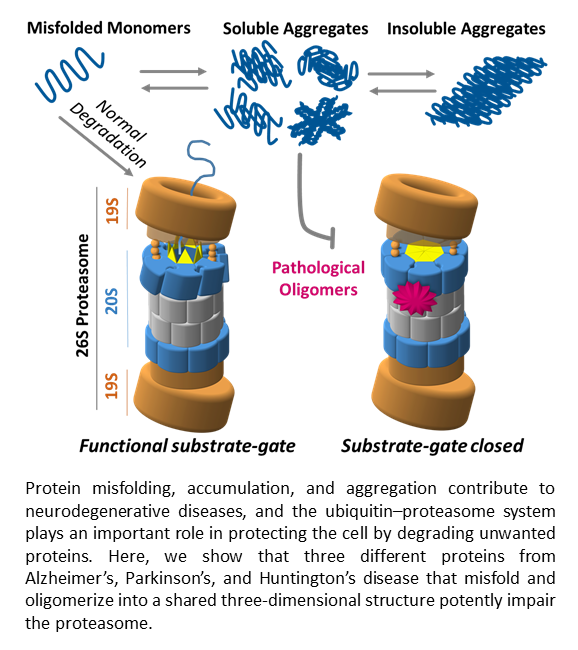
Proteasome and Neurodegenerative Disease
An important function of the proteasome is to degrade damaged or misfolded proteins in the cell. Most neurodegenerative diseases (e.g. Alzheimer’s, Huntington’s, Parkinson’s and Prion diseases) are characterized by accumulation of such neurotoxic proteins, and while causation for these diseases is elusive it is obvious that the cell’s only selective degradation machinery, the proteasome, must play an important role in their development. In fact it has been observed by many groups that such neurotoxic aggregates can negatively impact the ubiquitin-proteasome system. We are therefore trying to understand how toxic oligomers may inhibit or impair proteasome function, which could allow for a better understanding of the general mechanisms of these diseases and could lay the basis for future efforts to stimulate or restore proteasome function. Any opportunity to increase the functional capacity of the proteasome in specific ways could offer significant therapeutic benefits. Therefore, another interest in our lab is to design activators of protein degradation that might stimulate the degradation of toxic proteins involved in such neurodegenerative diseases.
Outreach Article: Regulators of protein degradation as potential treatments for neurodegenerative disease
Proteasome and Cancer
The 26S proteasome is the primary site for targeted protein degradation in mammalian cells and it is essential. An inhibitor of the proteasome, Velcade (Bortezomib, PS341) is now a widely used drug for the treatment of hematological cancers and is currently in more than 300 clinical trials. Because of the unpleasant side effects and emergence of resistance in many patients, there is appreciable interest for the development of new more specific inhibitors. While, Velcade inhibits proteasome function generally—by targeting its proteolytic sites—the proteasome actually contains more attractive sites for drug development that could block protein breakdown in more specific and targeted ways. One of our goals is to understand how we might develop new types of proteasome inhibitors whose mechanism and properties would differ markedly from such proteasome inhibitors now used in the clinic.
Several different types of proteasome regulatory complexes exist in mammalian cells that stimulate degradation of specific substrates. In addition, most of these complexes play important roles in cancer survival and progression, though their mechanisms are not understood. Our lab is interested in understanding how these cancer targets function to simulate protein degradation on a mechanistic level. These studies will hopefully lead us to a more general understanding of proteasome function in cancer, and in addition allow us to design new types of more specific proteasome inhibitors in the future.
Student opportunities
In this lab students will have the opportunities to learn a variety of biochemical, biophysical, structural, pharmacological, computational, molecular, and cell biological techniques. These skills are valuable for understanding and investigating enzyme function, drugs discovery, and the cell biology of cancer and neurodegenerative disease. Please feel free to contact me to learn more about what we do in our lab.
Grants and Research
Research Grants and Funding
NIH R01 GM107129 David Smith (PI) 07/2014-08/2023
Mechanisms regulating proteasomal substrate degradation $1,455,564
The goal of this grant is to understand how the mechanisms underlying how ATPase complexes recognize substrates and utilize the 20S proteasome to catalyze substrate degradation.
NIH R01 AG064188 David Smith (PI) 05/2020-04/2025
Proteasome function in Alzheimer's Disease $2,304,030
The goal of this grant is to demonstrating that the pathological oligomers associated with AD cause neuronal dysfunction, at least in part, by directly inhibiting the proteasome.
NIH R01 EY030050 Max Sokolov (PI) David Smith (Co-I) (5% effort) 05/2019-05/2023
Protein-unfolding chaperones for the treatment of blindness $1,500,000
The goal of this project is to develop a novel therapeutic application that utilizes a protein unfolding ATPase from archaea (PAN) for the treatment of inherited retinal dystrophies, including retinitis pigmentosa.
NIH R01 GM107129 David Smith (PI) 07/2014-08/2019
Mechanisms regulating proteasomal substrate degradation $1,425,000
The goal of this grant is to understand how the mechanisms underlying how ATPase complexes recognize substrates and utilize the 20S proteasome to catalyze substrate degradation.
International Myeloma Foundation David Smith (PI) 06/01/2012 - 05/30/2013
Specific Inhibitors of Ubiquitin-Dependent Proteasomal Degradation. $75,000
The goal of this project is to design and develop ubiquitin-dependent (26S) specific proteasome inhibitors to treat Multiple Myeloma.
Role: PI
Additional Info
PubMed Link: (Info can also be found under the Details Tab)
https://www.ncbi.nlm.nih.gov/myncbi/browse/collection/40498329/?sort=date&direction=descending