Contact
Positions
Professor Emeritus
- Organization:
- West Virginia University School of Medicine
- Department:
- Biochemistry and Molecular Medicine
- Classification:
- Unknown
Publications
Suchanek, A and Salati, L.M. (2015) "Construction and evaluation of an adenoviral vector for the liver-specific expression of the serine/arginine-rich splicing factor, SRSF3." Plasmid. 82:1-9
Cyphert, T.J., Suchanek, A.L., Griffith, B.N., and Salati, L.M. (2013) "Starvation activity inhibits splicing of glucose-6-phosphate dehydrogenase mRNA via a bifunctional ESE/ESS element bound by hnRNP K". Biochim. Biophys. Acta 1829:905-15.
Walsh, C.M.*, Suchanek, A.L.*, Cyphert, T.J., Kohan, A. B., Szeszel-Fedorowicz, W., and Salati, L.M. (2013) "Serine Arginine Splicing Factor 3 (SRSF3) is involved in enhanced splicing of glucose-6-phosphate dehydrogenase RNA in response to nutrients and hormones in liver". J. Biol. Chem. 288:2816-28. (*joint first authors)
Cyphert, H.A., Ge, X., Kohan, A.B., Salati, L.M., Zhang, Y., and Hillgartner, F.B. (2012) "Activation of the farnesoid X receptor induces hepatic expression and secretion of fibroblast growth factor 21". J. Biol. Chem. 287:25123-38.
Kohan AB, Qing Y, Cyphert HA, Tso P, Salati L.M. "Chylomicron remnants and nonesterified fatty acids differ in their ability to inhibit genes involved in lipogenesis in rats". J Nutr. 2011 Feb; 141(2):171-6. Epub 2010 Dec 15.
Griffith, B.N., Szeszel-Fedorowicz, W., Walsh, C.M., and Salati, L.M. (2006) "Identification of hnRNPs K, L and A2/B1 as candidate proteins involved in the nutritional regulation of mRNA splicing". Biochim. Biophys. A., 1759: 552-561.
Szeszel-Fedorowicz, W., Talukdar, I, Walsh, C.M., and Salati, L.M. (2006) "An exonic splicing silencer is involved in the regulated splicing of glucose-6-phosphate dehydrogenase mRNA". J. Biol. Chem., 281:34146-34158.
Research Program
Nutrient Control of Cellular Function and Metabolism
Research Interests
Nutrient Control of Cellular Function and Metabolism
Disease applications: Atherosclerosis, diabetes mellitus, metabolic syndrome, obesity
Basic science areas of research: Regulation of metabolism, Energy Homeostasis, Hormone action, Regulation of gene expression, mRNA processing, Protein-nucleic acid interactions, Nutritional biochemistry, Hepatocyte function
The long-term goal of the laboratory is to understand the molecular details by which fatty acids regulate cellular functions. Currently the laboratory is approaching this from two complementary and ultimately intersecting approaches:
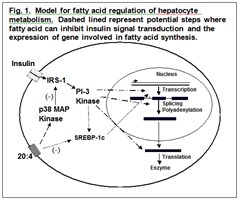
Inhibition of insulin signal transduction by fatty acids. Insulin is a major hormonal signal is the intact animal. When humans and other animals consume a diet rich in carbohydrate, blood insulin levels increase and the flux through metabolic pathways favor the storage of excess carbohydrate as glycogen and as the fat, triacylglycerol. The liver is a major site for the conversion of carbohydrate to fat. Flux through this synthetic pathway is decreased when polyunsaturated fat is included in the diet. Our data provides evidence that the decrease in flux is caused by a decrease in insulin signal transduction within the liver. We are defining the pathways by which fatty acids interfere with insulin signal transduction and ultimately regulate the amounts of the enzymes that synthesize fat (see Fig. 1). When the amount of fat in the diet is low the regulation of fat synthesis is reversible but when the amount of fat in the diet increases cellular metabolism is irreversibly altered. It is these latter events that lead to metabolic syndrome. Our goal is to define the steps involved in fatty acid signaling and to distinguish between the normal reversible regulatory mechanisms from the long-term adaptations that occur during obesity and diabetes.
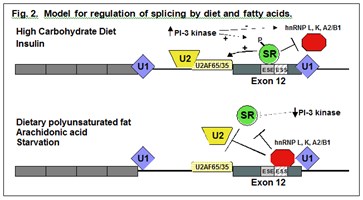
Molecular mechanism by which polyunsaturated fats inhibit gene expression. We have chosen to study the mechanism by which fatty acids inhibit gene expression using glucose-6-phosphate dehydrogenase (G6PD) as our model gene. G6PD provides the reducing equivalents for the synthesis of fatty acids and its activity correlates both with the capacity of the liver to synthesize fat and with the amount of circulating blood lipids. My laboratory has discovered a novel mechanism by which polyunsaturated fatty acids and in particular arachidonic acid can regulate gene expression: decreasing the rate of pre-mRNA splicing. This mechanism accounts most of the regulation of G6PD expression and does so in the absence of changes in G6PD gene transcription. We are analyzing molecular basis for the regulation of mRNA splicing by nutrients. These experiments have defined a splicing regulatory element that contains both an enhancer element (ESE) and an inhibitory or silencing element (ESS). We have further identified candidate splicing regulatory proteins involved in the nutritional regulation of mRNA splicing (see Fig. 2). SR proteins bind to the ESE an increase the rate of splicing; while hnRNPs L, K and A2/B1 bind the ESS and inhibit splicing. Our ultimate goal is to determine the mechanisms by which the activities of these proteins are altered by dietary components.
Research Support: This research has been supported by the National Institutes of Health, the American Heart Association, and the American Cancer Society