Contact
Positions
Res. Assistant Professor
- Organization:
- West Virginia University School of Medicine
- Department:
- Biochemistry and Molecular Medicine
- Classification:
- Faculty
Member
- Organization:
- West Virginia University WVU Cancer Institute
- Department:
- WVU Cancer Institute Research Programs
- Classification:
- Faculty
Education
- MS, Moscow State University, 1994
- PhD, Engelhardt Institute of Molecular Biology, Russian Academy of Sciences, 1999
Publications
Ye Q, Hickey J, Summers K, Falatovich B, Gencheva M, Eubank TD, Ivanov AV*, Guo NL*. Multi-Omics Immune Interaction Networks in Lung Cancer Tumorigenesis, Proliferation, and Survival. International Journal of Molecular Sciences. 2022, 23(23):14978. PMID: 36499305, PMCID: PMC9738413, DOI:10.3390/ijms232314978
Ye Q, Falatovich B, Singh S, Ivanov AV, Eubank TD, and Guo NL. A Multi-Omics Network of a Seven-Gene Prognostic Signature for Non-Small Cell Lung Cancer. International Journal of Molecular Sciences. 2022, 23(1):219. PMID: 35008645, PMCID: PMC8745553, DOI: 10.3390/ijms23010219
Ye Q, Mohamed R, Dukhlallah D, Gencheva M, Hu G, Pearce MC, Kolluri SK, Marsh CB, Eubank TD, Ivanov AV, Guo NL. Molecular Analysis of ZNF71 KRAB in Non-Small-Cell Lung Cancer International Journal of Molecular Sciences. 2021, 22(7):3752. PMID: 33916522, PMCID: PMC8038441, DOI: 10.3390/ijms22073752
Addison JB, Voronkova MA, Fugett JH, Lin CC, Linville NC, Trinh B, Livengood RH, Smolkin MB, Schaller MD, Ruppert JM, Pugacheva EN, Creighton CJ, and Ivanov AV. Functional Hierarchy and Cooperation of EMT Master Transcription Factors in Breast Cancer Metastasis Molecular Cancer Research, 2021, 19(5):784-798; Epub 2021 Jan 26. PMID: 33500360. DOI: 10.1158/1541-7786.MCR-20-0532
Zhao H, Martin E, Shah Neal, Ivanov A, Ruppert JM, Lockman P, and Agazie YM. Conditional knockout of SHP2 in ErbB2 transgenic mice or inhibition in HER2-amplified breast cancer cell lines blocks oncogene expression and tumorigenesis Oncogene. 2018, 38(13):2275-2290. PMID: 30467378, PMCID: PMC6440805.
Park J, Schlederer M, Schreiber M, Ice R, Merkel O, Bilban M, Hofbauer S, Kim S, Addison J, Zou J, Ji C, Bunting ST, Wang Z, Shoham M, Huang G, Bago-Horvath Z, Gibson LF, Rojanasakul Y, Remick S, Ivanov A, Pugacheva E, Bunting KD, Moriggl R, Kenner L, Tse W. AF1q is a novel TCF7 co-factor which activates CD44 and promotes breast cancer metastasis Oncotarget. 2015 Aug 21;6(24):20697-710. PMID: 26079538, PMCID: PMC4653036.
Loskutov YV, Kozyulina PY, Kozyreva VK, Ice RJ, Jones BC, Roston TJ, Smolkin MB, Ivanov AV, Wysolmerski RB, and Pugacheva EN. NEDD9/Arf6-dependent endocytic trafficking of matrix metalloproteinase 14: a novel mechanism for blocking mesenchymal cell invasion and metastasis of breast cancer Oncogene. 2015 Jul;34(28):3662-75. Epub 2014 Sep 22. PMCID: PMC4369482.
Addison JB, Koontz C, Fugett JH, Creighton CJ, Chen D, Farrugia MK, Padon RR, Voronkova MA, McLaughlin SL, Livengood RH, Lin CC, Ruppert JM, Pugacheva EN and Ivanov AV. KAP1 Promotes Proliferation and Metastatic Progression of Breast Cancer Cells Cancer Research. 2015 Jan 15;75(2):344-55. Epub 2014 Nov 24. PMCID: PMC4297582.
McLaughlin SL, Ice RJ, Rajulapati A, Kozyulina PY, Livengood RH, Kozyreva VK, Loskutov YV, Culp MV, Weed SA, Ivanov AV, Pugacheva EN. NEDD9 Depletion Leads to MMP14 Inactivation by TIMP2 and Prevents Invasion and Metastasis Molecular Cancer Research. 2014 Jan; 12(1):69-81. Epub 2013 Nov 7. PMCID: PMC3946989.
Pacurari M, Addison JB, Bondalapati N, Wan YW, Luo D, Qian Y, Castranova V, Ivanov AV*, Guo NL*. The microRNA-200 family targets multiple non-small cell lung cancer prognostic markers in H1299 cells and BEAS-2B cells International Journal of Oncology. 2013 Aug; 43(2):548-60. Epub 2013 May 27. PMCID: PMC3775564.
Ice RJ, McLaughlin SL, Livengood RH, Culp M, Eddy ER, Ivanov AV, Pugacheva EN. NEDD9 Depletion Destabilizes Aurora A Kinase and Heightens the Efficacy of Aurora A Inhibitors: Implications for Treatment of Metastatic Solid Tumors Cancer Research. 2013 May 15; 73(10):3168-80. Epub 2013 Mar 28. PMCID: PMC3667743.
Cieply B, Riley P, Pifer P, Widmeyer J, Addison JB, Ivanov AV, Denvir J, Frisch SM. Suppression of the Epithelial–Mesenchymal Transition by Grainyhead-like-2 Cancer Research, 2012 May 1; 72(9):2440-53. Epub 2012 Feb 29. PMCID: PMC3342427.
White D, Rafalska-Metcalf IU, Ivanov AV, Corsinotti A, Peng H, Lee SC, Trono D, Janicki SM, Rauscher FJ 3rd. The ATM Substrate KAP1 Controls DNA Repair in Heterochromatin: Regulation by HP1 Proteins and Serine 473/824 Phosphorylation Molecular Cancer Research, 2012 Mar; 10(3):401-14. Epub 2011 Dec 28. PMID: 22205726 PMCID: PMC4894472.
Lin CC, Liu LZ, Addison JB, Ivanov AV, Ruppert JM. A KLF4–miRNA-206 Autoregulatory Feedback Loop Can Promote or Inhibit Protein Translation Depending upon Cell Context Molecular and Cellular Biology, 2011 Jun; 31(12):2513-27. Epub 2011 Apr 25. PMCID: PMC3133414.
Iyengar S, Ivanov AV, Jin VX, Rauscher FJ, Farnham PJ. Functional Analysis of KAP1 Genomic Recruitment Molecular and Cellular Biology. 2011 May; 31(9):1833-47. Epub 2011 Feb 22. PMCID: PMC3133220.
Peng H, Ivanov AV, Oh HJ, Lau YF, Rauscher FJ. Epigenetic Gene Silencing by the SRY Protein Is Mediated by a KRAB-O Protein That Recruits the KAP1 Co-repressor Machinery Journal of Biological Chemistry. 2009 Dec 18;284(51):35670-80. PMCID: PMC2790998.
Zeng L, Yap KL, Ivanov AV, Wang X, Mujtaba S, Plotnikova O, Rauscher FJ III, Zhou M-M. Structural insights into human KAP1 PHD finger–bromodomain and its role in gene silencing Nature Structural and Molecular Biology. 2008 Jun;15(6):626-33. Epub 2008 May 18. PMCID: PMC3331790.
Sarma K, Margueron R, Ivanov A, Pirotta V and Reinberg D. Ezh2 Requires PHF1 To Efficiently Catalyze H3 Lysine 27 Trimethylation In Vivo Molecular and Cellular Biology. 2008 Apr;28(8):2718-31. Epub 2008 Feb 19. PMCID: PMC2293112.
Ivanov A, Peng H, Yurchenko V, Negorev D, Fredericks W, Maul G, Sadofsky M, Zhou M-M, Rauscher F, III. PHD Domain-Mediated E3 Ligase Activity Directs Intramolecular Sumoylation of an Adjacent Bromodomain Required for Gene Silencing Molecular Cell. 2007 Dec 14;28(5):823-37. PMCID: PMC4348069.
Research Program
Alexander B. Osborn Hematopoietic Malignancy and Transplantation Program
Research Interests
Highlighted Project 1:
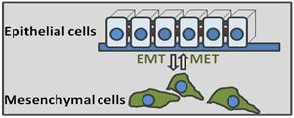
Cancer is a disease of gene expression. Through studies of breast and lung cancer we are working to understand general principles of gene misregulation in cancer cells with particular emphasis on gene transcription and microRNAs. Vast majority of human tumors are of epithelial origin, e.g. they derive from cells normally highly organized in specialized epithelial layers. At the same time, most cancer-related deaths occur due to tumor recurrence and spread to distant organs (metastasis), which are tightly linked to acquisition by cancer cells of mesenchymal properties such as increased motility, invasion and resistance to chemotherapy. The metastasis stage of cancer is associated with the epithelial-to-mesenchymal transition (EMT). Normally activated only during early embryonic development, the EMT program is highjacked by cancer cells during evolution of individual tumors. EMT is activated by a handful or transcription factors referred to as the EMT master regulators, such as Snail and ZEB.
The goals of our research are to identify transcriptional network involved in activation of EMT during cancer metastasis. This knowledge will help to develop future therapeutic approaches in treating cancer and prevention of metastasis.
Relevant Publications:
- Addison JB, Voronkova MA, Fugett JH, Lin CC, Linville NC, Trinh B, Livengood RH, Smolkin MB, Schaller MD, Ruppert JM, Pugacheva EN, Creighton CJ, and Ivanov AV. Functional Hierarchy and Cooperation of EMT Master Transcription Factors in Breast Cancer Metastasis Molecular Cancer Research, Epub 2021 Jan 26.
- Cieply B, Riley P, Pifer P, Widmeyer J, Addison JB, Ivanov AV, Denvir J, Frisch SM. Suppression of the Epithelial–Mesenchymal Transition by Grainyhead-like-2 Cancer Research, 2012 May 1; 72(9):2440-53.
- Pacurari M, Addison JB, Bondalapati N, Wan YW, Luo D, Qian Y, Castranova V, Ivanov AV*, Guo NL*. The microRNA-200 family targets multiple non-small cell lung cancer prognostic markers in H1299 cells and BEAS-2B cells International Journal of Oncology. 2013 Aug; 43(2):548-60.
Highlighted Project 2:
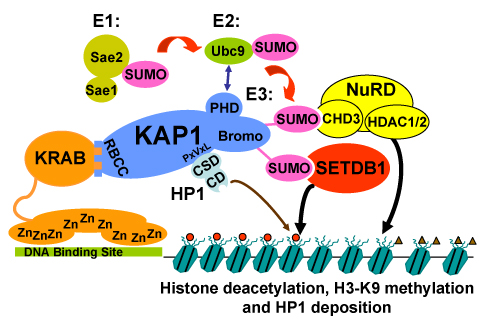
Eukaryotic genomes are, in general, in a default state of repression, where the vast majority of genes are turned off or silenced. This repression is accomplished largely through packaging DNA into tightly coiled DNA-protein fibers called chromatin by association with histones and other proteins. This means that chromatin must be uncoiled before allowing genes to be accessed by proteins that mediate transcription. One mechanism that regulates chromatin structure is the attachment of chemical groups to the histones. It is now becoming clear that a diverse array of enzymes modify histones and place a range of various chemical modifications, including acetylation, phosphorylation, methylation, and sumoylation. One appealing idea is that the pattern and identity of histone modifications constitute a "code" for specific processes, such as transcription. The histone code is written in proteins, not DNA, it can be heritable and constitutes the basis of epigenetics which is increasingly recognized as an important factor in cancer development.
The main system for studying the mechanisms of transcriptional repression in the lab is KRAB domain containing transcription factors. The KRAB-zinc finger (KRAB-ZNF) superfamily of DNA-binding transcriptional repressors is the largest family of gene silencers encoded in the human genome: Estimates are that of the >700 Cys2-His2 class zinc finger genes, more than 350 contain the highly conserved ~75 amino acid KRAB repression domain. KRAB-ZNF mediated transcriptional silencing requires a direct interaction with an obligate co-repressor KAP1 (TRIM28), which serves as a scaffolding protein for recruitment of repression machinery. KAP1 plays important roles in embryonic development, stem cell self-renewal, chromatin organization, and the DNA damage response, acting as an essential corepressor for KRAB-ZNFs. Given the relevance of developmental cell fate regulators and stem cell pluripotency to cancer pathogenesis, understanding how KAP1 functions in cancer cells might be critical for developing future therapeutic strategies.
We recently showed that KAP1 promotes breast cancer cell proliferation and metastasis. The goals of our research are to elucidate the molecular mechanisms by which KAP1 contributes to cancer progression.
Relevant Publications:
- ZINC FINGER LINKER (ZnFL) ANTIBODY - Patent US-2016200829-A1 - We have shown that the ZnFL antibody specifically detects multiple ZNF transcription factors overexpressed in cancer cells, suggesting that they could be used as potential diagnostic markers. Please inquire about licensing of the ZnFL antibody.
- Anti-Zinc Finger Linker (ZnFL) Antibody - commercially available from EMD/Millipore/Sigma cat.#ABE1941
- Addison JB, Koontz C, Fugett JH, Creighton CJ, Chen D, Farrugia MK, Padon RR, Voronkova MA, McLaughlin SL, Livengood RH, Lin CC, Ruppert JM, Pugacheva EN and Ivanov AV. KAP1 Promotes Proliferation and Metastatic Progression of Breast Cancer Cells Cancer Research. 2015 Jan 15;75(2):344-55.
- White D, Rafalska-Metcalf IU, Ivanov AV, Corsinotti A, Peng H, Lee SC, Trono D, Janicki SM, Rauscher FJ 3rd. The ATM Substrate KAP1 Controls DNA Repair in Heterochromatin: Regulation by HP1 Proteins and Serine 473/824 Phosphorylation Molecular Cancer Research, 2012 Mar; 10(3):401-14.
- Ivanov A, Peng H, Yurchenko V, Negorev D, Fredericks W, Maul G, Sadofsky M, Zhou M-M, Rauscher F, III. PHD Domain-Mediated E3 Ligase Activity Directs Intramolecular Sumoylation of an Adjacent Bromodomain Required for Gene Silencing Molecular Cell. 2007 Dec 14;28(5):823-37.